- Title
-
Semaphorin 3f and post-embryonic regulation of retinal progenitors
- Authors
- Kalifa, A., Hehr, C.L., Shewchuk, K.L., Mori-Kreiner, R., Standing, S., Halabi, R., McFarlane, S.
- Source
- Full text @ PLoS Genet.
|
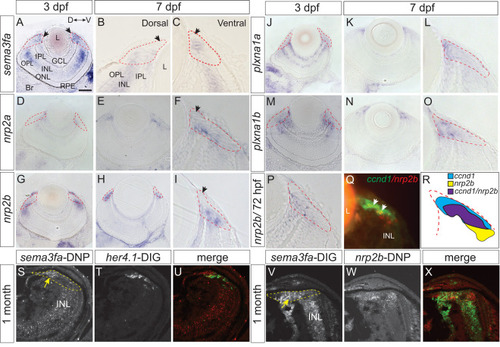
Expression of mRNAs for Sema3fa and its receptors in the larval and juvenile CMZ. Plastic sections through the eyes of WM ISH of 3 (A, D, G, J, M, P) and 7 (B, C, E, F, H, I, K, L, N, O) dpf larvae for |
|
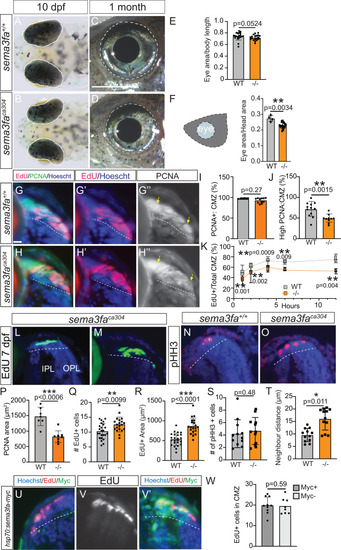
Larval eye size and CMZ proliferating progenitors impacted in the |
|
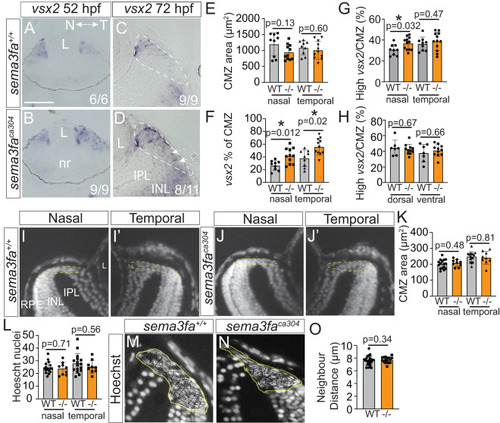
Gross CMZ development and anatomy unimpacted by Sema3fa loss. |
|
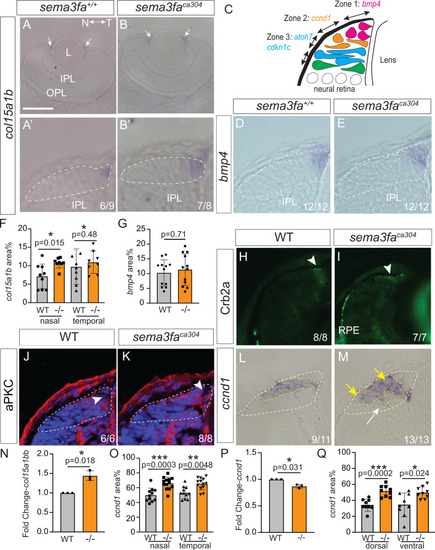
Disruption of retinal progenitor cell domain with loss of Sema3fa. |
|
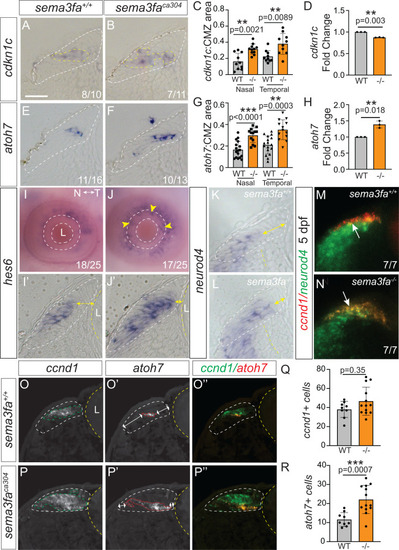
Disrupted organization of committed CMZ progenitors in the absence of Sema3fa. WM ISH for Zone 3 markers shown in 7 µm plastic sections (A, B, E, F, I′, J′, K, L), z-stack of confocal optical sections |
|
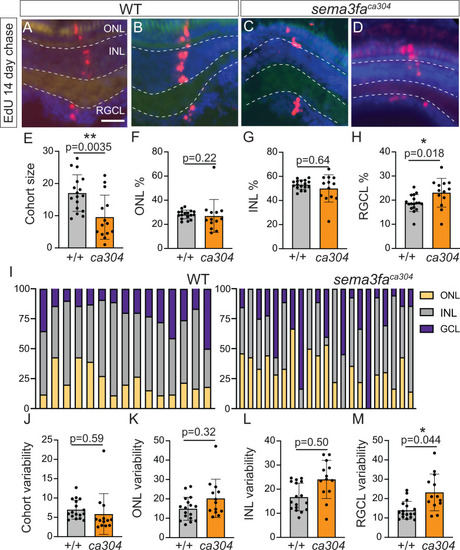
Altered size and cell composition of CMZ-derived radial cohorts with loss of Sema3fa. EdU labelling of retinas of 20 dpf zebrafish larvae incubated for 4 hrs with EdU at 5 dpf. |
|
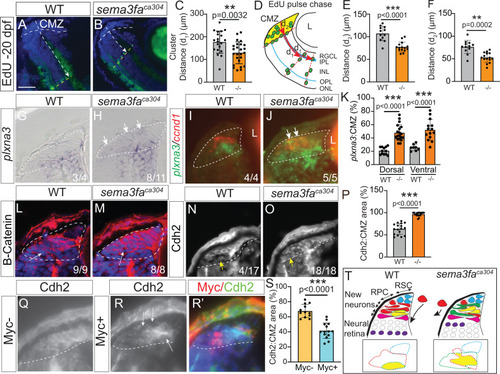
Sema3fa may promote smooth movement of progenitors through CMZ by downregulating cell adhesion. |