- Title
-
CDKL1 variants affecting ciliary formation predispose to thoracic aortic aneurysm and dissection
- Authors
- Nauth, T., Philipp, M., Renner, S., Burkhalter, M.D., Schüler, H., Saygi, C., Händler, K., Siebels, B., Busch, A., Mair, T., Rickassel, V., Deden, S., Hoffer, K., Olfe, J., Mir, T.S., von Kodolitsch, Y., Girdauskas, E., Rybczynski, M., Kriegs, M., Voß, H., Sauvigny, T., Spielmann, M., Alawi, M., Krasemann, S., Kubisch, C., Demal, T.J., Rosenberger, G.
- Source
- Full text @ Journal of Clin. Invest.
|
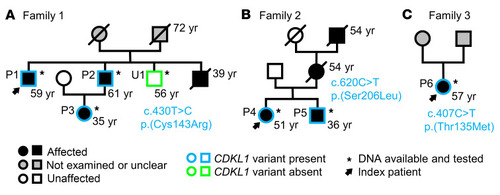
Family pedigrees. The respective identified |
|
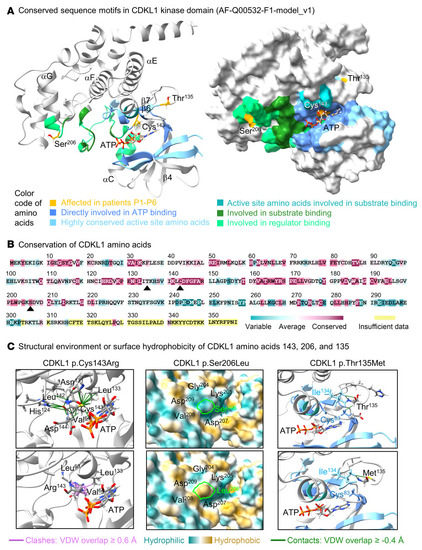
Structural and physicochemical impact of CDKL1 amino acid changes. ( |
|
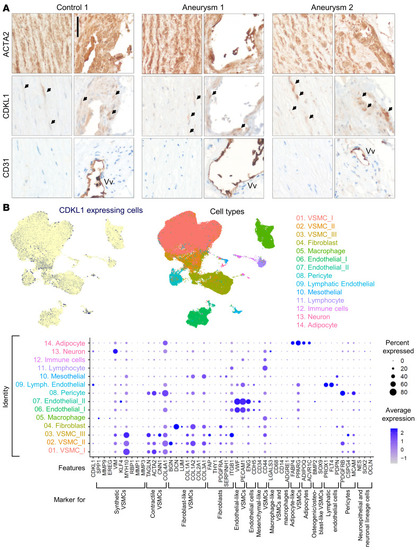
CDKL1 is expressed in VSMCs of normal and diseased aortic tissue. ( |
|
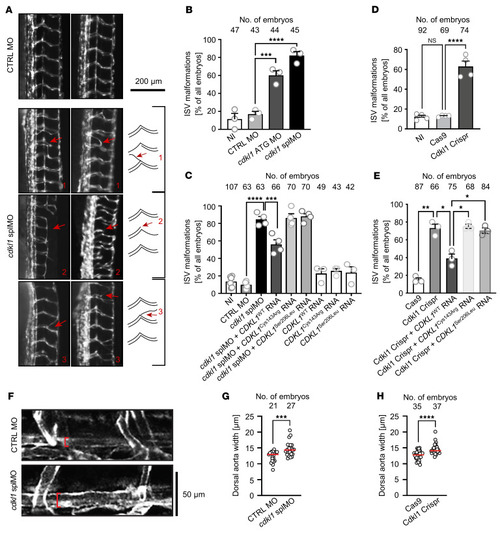
ISV malformation and aortic dilation in zebrafish ( |
|
( |
|
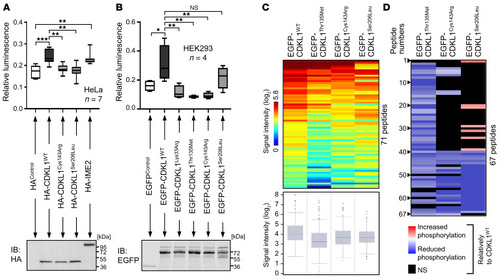
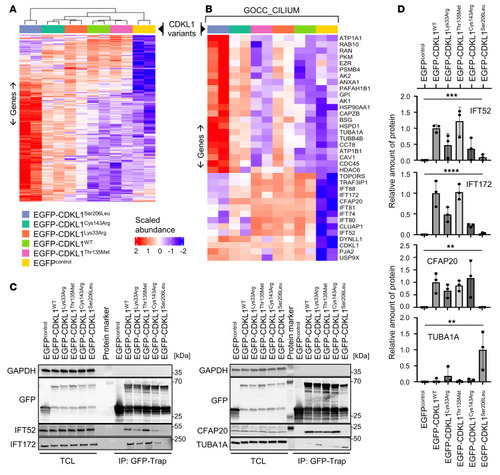
Disease-associated CDKL1 variants show different protein-protein interaction profiles. EGFP-tagged CDKL1 protein variants were expressed in HEK293T cells, purified by GFP-Trap, and subjected to liquid chromatography–tandem mass spectrometry analysis. Experimental duplicates were performed for all conditions. Protein abundances were normalized based on GFP protein peptide abundances. ( |
|
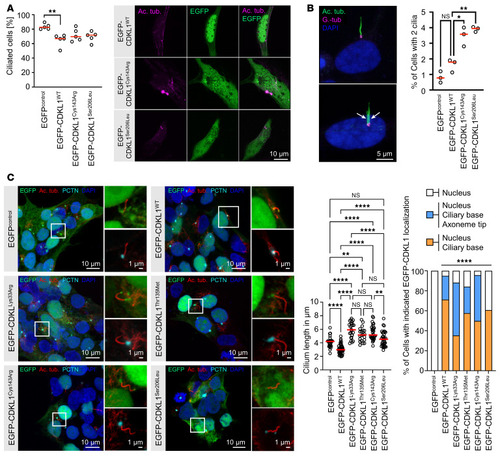
Consequences of CDKL1 variants for ciliary biology. ( |
|
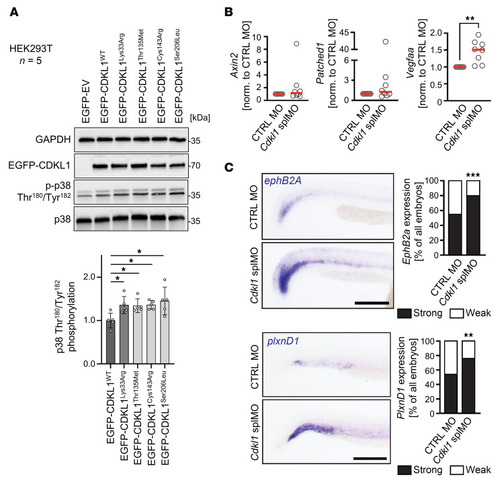
CDKL1 function in cell signaling. ( |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. EXPRESSION / LABELING:
|