Fig. 4
- ID
- ZDB-FIG-250529-37
- Publication
- Zhang et al., 2024 - Znf706 regulates germ plasm assembly and primordial germ cell development in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
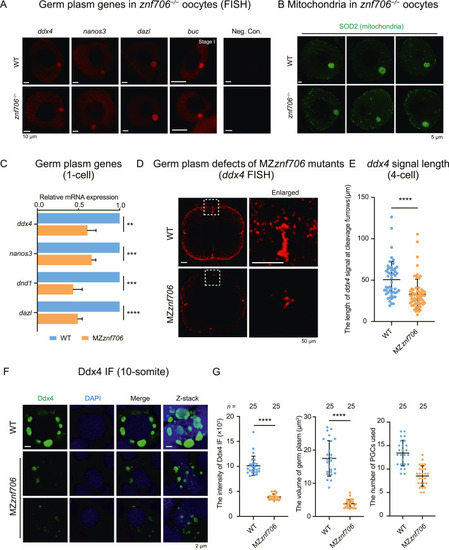
Znf706 is required for normal germ plasm assembly in PGCs. A: FISH of germ plasm genes (ddx4, nanos3, dazl, and buc) in WT and znf706−/− oocytes at stage I. B: Immunostaining of SOD2 showing the assembly of mitochondria in WT and znf706−/− stage I oocytes. C: Bar plot showing relative expression levels of germ plasm genes (ddx4, nanos3, dnd1, and dazl) in WT and MZznf706 embryos at 1-cell stage. D: FISH of ddx4 showing germ plasm at the cleavage furrows of WT and MZznf706 embryos at 4-cell stage. The dashed boxed areas were enlarged for better viewing. E: Scatter plot showing the length of ddx4 signals (indicating germ plasm) at cleavage furrows (D). Each dot in the scatter plot indicated ddx4 signal in one cleavage furrow in WT and MZznf706 embryos. The total numbers of dots are 51 in WT and 65 in MZznf706, respectively. F: Immunostaining of Ddx4 showing the distribution of germ plasm in WT and MZznf706 PGCs at 10-somite stage. G: Scatter plots showing the intensity (left), the volume of germ plasm granule (middle) of Ddx4 signals in PGCs, and the number of PGCs used for calculation (right) in WT and MZznf706 embryos at 10-somite stage (F). Twenty-five embryos in each group were analyzed. Scales bars, 10 μm (A); 5 μm (B); 50 μm (D); 2 μm (F). ∗∗∗∗, P < 0.0001; ∗∗∗, P < 0.001; ∗∗, P < 0.01. FISH, fluorescence in situ hybridization; MZ, maternal-zygotic; WT, wild-type; Neg. Con., negative control without probe. |
Reprinted from Journal of genetics and genomics = Yi chuan xue bao, 52, Zhang, W., Li, Y., Li, H., Liu, X., Zheng, T., Li, G., Liu, B., Lv, T., Wei, Z., Xing, C., Jia, S., Meng, A., Wu, X., Znf706 regulates germ plasm assembly and primordial germ cell development in zebrafish, 666-679, Copyright (2024) with permission from Elsevier. Full text @ J. Genet. Genomics