- Title
-
Disruption of grin2A, an epilepsy-associated gene, produces altered spontaneous swim behavior in zebrafish
- Authors
- Abramova, V., Tomovic, E., Kysilov, B., Korinek, M., Dobrovolski, M., Hrcka Krausova, B., Fili, K., Elzahraa S Abdel Rahman, F., Bozikova, P., Cerny, J., Smejkalova, T., Balik, A., Vyklicky, L.
- Source
- Full text @ J. Neurosci.
|
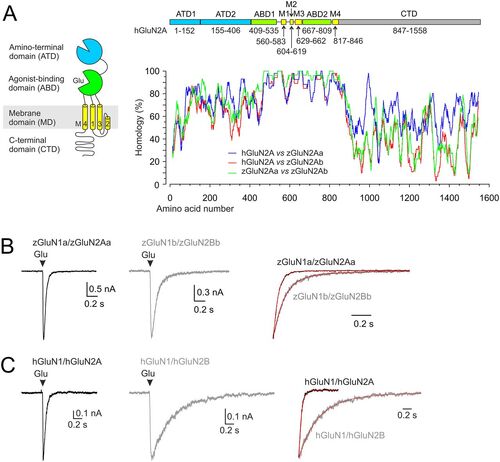
Protein sequence homology and functional properties of recombinant zebrafish NMDARs. A, Diagram of the domains comprising the hGluN2A subunit: the extracellular ATD and ABD; the membrane TMD with transmembrane segments M1, M3, M4, and the M2 pore loop forming the ion channel; and the intracellular CTD. A graph showing the amino acid alignment between hGluN2A (NP_001127879.1) and each of the zebrafish GluN2A paralogs zGluN2Aa and zGluN2Ab (XP_021329529.1, XP_009304490.1). Values indicate the percentage of homology calculated as a relative BLOSUM62 substitution matrix score between the ClustalW alignment of the indicated subunits. Each position in the alignment was scored by 100×2BLOSUM62(ri,ci)−BLOSUM62(ri,ri) with ri and ci corresponding to ith amino acid in the “reference” and “compared” sequence, respectively. A running average of 25 amino acid segments was then used for the plotting. Above the graph is the linear representation of the hGluN2A subunit. The colors correspond to the diagram, with the ATD shown in blue, the ABD in green, the TMD in yellow, and the CTD in gray. The amino acid numbers that make up each domain are indicated. B, C, Representative current recordings in HEK293T cells expressing zGluN1a/zGluN2Aa or zGluN1b/zGluN2Bb receptors (B) or hGluN1/hGluN2A or hGluN1/GluN2B receptors (C), with responses evoked by a brief (10–15 ms) application of 1 mM glutamate in the continuous presence of 100 µM glycine. The responses normalized to the peak amplitude are shown on the right (red lines indicate the double exponential function fit to the data, τw = 36.2 ms for zGluN1a/zGluN2Aa and τw = 118.9 ms for zGluN1b/zGluN2Bb; τw = 58.8 ms for hGluN1/hGluN2A and τw = 510 ms for hGluN1/hGluN2B). |
|
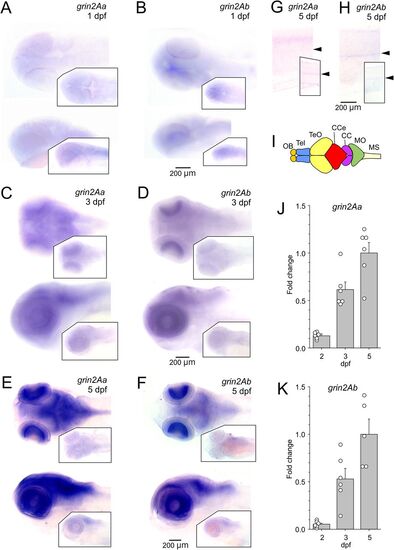
Expression of grin2Aa and grin2Ab in the zebrafish nervous system. Whole-mount ISH of grin2Aa and grin2Ab at 1 dpf (A, B), 3 dpf (C, D), and 5 dpf (E, F) showing dorsal (top) and lateral (bottom) views. A, C, E, Insets, sense probes of grin2Aa. B, D, F, Insets, sense probes of grin2Ab. G, H, Lateral view of the trunk at 5 dpf for grin2Aa (G) and grin2Ab (H). The position of the spinal cord is indicated by arrows. Insets, Sense probes of grin2Aa and grin2Ab. I, Simplified representation of the zebrafish central nervous system and its major structures. Adapted from Wullimann et al. (1996); CC, crista cerebellaris; CCe, corpus cerebelli; MO, medulla oblongata; MS, medulla spinalis; OB, olfactory bulb; Tel, telencephalon; TeO, tectum opticum. J, K, Relative expression levels of grin2Aa and grin2Ab mRNA at 2 dpf (n = 6; 6), 3 dpf (n = 6; 6), and 5 dpf (n = 6; 5) in the head of zebrafish larvae analyzed using RT-qPCR. Results are normalized to the corresponding gene expression at 5 dpf. For mRNA expression levels of grin1 and grin2B paralogs in the zebrafish nervous system, see Extended Data Figure 2-1. |
|
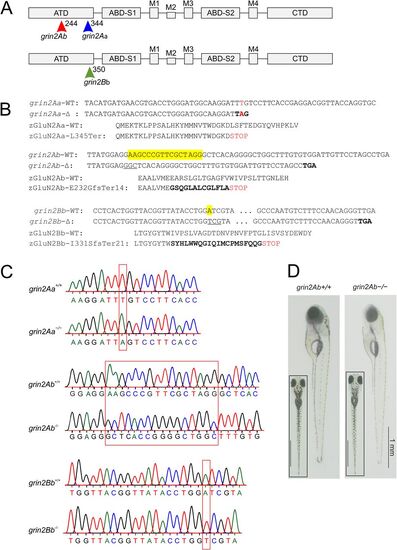
Locations of zGluN2 mutations resulting in a premature stop codon and protein truncation. A, Schematic representation of the domain structure of a GluN2 subunit. It consists of four domains: the extracellular ATD and S1 and S2 lobes of the ABD; the membrane TMD with transmembrane segments M1, M3, M4, and M2 pore loop forming the ion channel; and the intracellular CTD. In the ATD, arrows indicate the position of grin2Aa mutation (sa14573) and gRNA target sites for grin2Ab and grin2Bb. B, The nucleotide and amino acid sequence alignment of the portion of the ATD of zGluN2Aa wild-type (grin2Aa-WT; zGluN2Aa-WT) and the mutant strain (grin2Aa-Δ; zGluN2Aa-L345Ter; top), zGluN2Ab wild-type (grin2Ab-WT; zGluN2Ab-WT) and the mutant strain (grin2Ab-Δ; zGluN2Ab-E232GfsTer14; middle), and zGluN2Bb wild-type (grin2Bb-WT; zGluN2Bb-WT) and the mutant strain (grin2Bb-Δ; zGluN2Bb-I331SfsTer21; bottom). C, Chromatograms of Sanger sequencing of PCR amplicons of genomic DNA from grin2Aa+/+ and grin2Aa−/− (top), grin2Ab+/+ and grin2Ab−/− (middle), and grin2Bb+/+ and grin2Bb−/− (bottom) larvae. D, Lateral and dorsal (inset) images of representative 6 dpf wild-type (grin2Ab+/+) and grin2Ab−/− larvae. For viability of larvae with mono- and biallelic deletion of grin2Aa, grin2Ab, or both genes at 6 dpf, see Extended Data Figure 3-1. For growth of the grin2Aa−/−, grin2Ab−/−, and grin2A−/− mutant fish during development, see Extended Data Figure 3-2. PHENOTYPE:
|
|
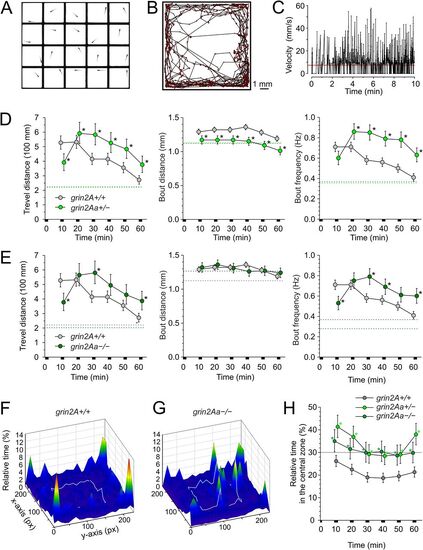
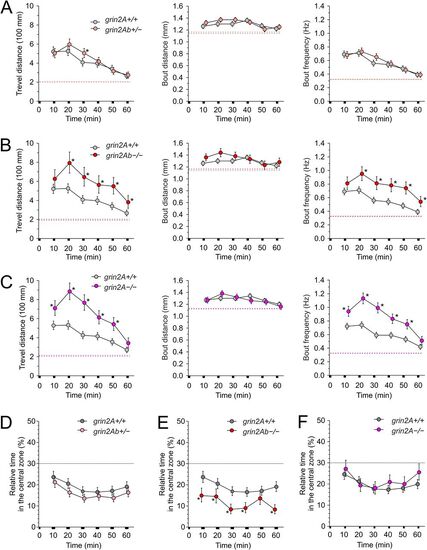
Effects of grin2Aa deletion on larval swimming behavior and thigmotaxis. A, Photograph of a multiwell chamber used to simultaneously assess the spontaneous swimming behavior of 20 larvae (6 dpf). Larvae in the experiment were progeny of grin2Aa+/− intercrosses with the genotype determined after the behavioral experiment. B, The graph shows the sample trajectory of a grin2Aa+/+ larva as assessed during the first 10 min interval. Data points were acquired at 50 Hz. C, The graph shows the velocity of a grin2A+/+ larva assessed at 20 ms intervals. The red line indicates the velocity threshold for locomotor activity detection (see Materials and Methods). The MTD, MBD, and MBF of grin2Aa+/− and grin2A+/+ larvae (D) and grin2Aa−/− and grin2A+/+ larvae (E) are compared. Dashed lines correspond to mean values of the measured parameters at steady state, 2–3 h after the placement of larvae into the experimental chamber. For steady-state swimming parameters assessed 2–3 h after the larvae were placed in the experimental chambers, see Extended Data Figure 4-1. Representative heat maps for the relative time spent by grin2A+/+ (F) and grin2Aa−/− (G) larvae in 0.454 × 0.454 mm area bins (10 × 10 pixels) over 20 min of recording (30–50 min), indicated by color coding. The central zone (5.5 × 5.5 mm) used to assess thigmotaxis is outlined by a gray line. H, Comparison of thigmotaxis measured as the relative time spent by grin2A+/+, grin2Aa+/−, and grin2Aa−/− larvae in the central zone. The analysis was performed on the same larvae used to study locomotion. The dotted line indicates the level of randomness (30%). Data are expressed as mean ± 95% confidence interval of travel distance, bout distance, and bout frequency (D, E), and the relative time spent by larvae in the central zone (H) in episodes lasting 10 min; * indicates significant differences in genotype as assessed by ANOVA followed by the LSD post hoc test (grin2A+/+, n = 582; grin2Aa+/−, n = 429; grin2Aa−/−, n = 216). grin2Aa+/− versus grin2A+/+, travel distance genotype, p < 0.001; time, p < 0.001; bout distance genotype, p < 0.001; time, p < 0.001; bout frequency, genotype, p < 0.001; time, p < 0.001; and relative time larvae spent in the central zone genotype, p < 0.001; time, p < 0.001. grin2Aa−/− versus grin2A+/+, travel distance genotype, p = 0.0056; time, p < 0.001; bout distance genotype, p = 0.81; time p = 0.013; bout frequency, genotype, p = 0.001; time, p < 0.001; and relative time spent by larvae in the central zone genotype, p < 0.001; time, p = 0.013. PHENOTYPE:
|
|
Effects of grin2Ab deletion on larval swimming behavior and thigmotaxis. Comparison of MTD, MBD, and MBF of grin2Ab+/− (A), grin2Ab−/− (B), and grin2A−/− (C) larvae compared with grin2A+/+ larvae (6 dpf). Mean steady-state values of the locomotion parameters examined 2–3 h after the placement of larvae in the experimental wells are shown as dashed lines. For steady-state swimming parameters assessed 2–3 h after the larvae were placed in the experimental chambers, see Extended Data Figure 4-1. D, Comparison of thigmotaxis measured as the relative time spent in the central zone by grin2Ab+/−, grin2Ab−/−, and grin2A−/− larvae compared with grin2A+/+ larvae. The analysis of thigmotaxis was performed on the same larvae used to study locomotion. The dotted line indicates the level of randomness (30%). Data are expressed as mean ± 95% confidence interval of travel distance, bout distance, and bout frequency (A–C), and the relative time spent by larvae in the central zone (D) measured in 10 min intervals; * indicates significant differences in the genotype as assessed by ANOVA followed by LSD post hoc test (grin2A+/+, n = 584; grin2Ab+/−, n = 542; grin2Ab−/−, n = 197; grin2A−/−, n = 270). grin2Ab+/− versus grin2A+/+, travel distance genotype, p = 0.26; time, p < 0.001; bout distance genotype, p = 0.062; time, p < 0.001; bout frequency genotype, p = 0.55; time, p < 0.001; the relative time larvae spent in the central zone genotype, p = 0.001; time, p < 0.001. grin2Ab−/− versus grin2A+/+, travel distance genotype, p < 0.001; time, p < 0.001; bout distance genotype, p = 0.051; time, p = 0.003; bout frequency genotype, p < 0.001; time, p < 0.001; the relative time larvae spent in the central zone genotype, p < 0.001; time, p = 0.001. grin2A−/− versus grin2A+/+, travel distance genotype, p < 0.001; time, p < 0.001; bout distance genotype, p = 0.50; time, p < 0.001; bout frequency genotype, p < 0.001; time, p < 0.001; and the relative time larvae spent in the central zone genotype, p = 0.10; time, p < 0.001. PHENOTYPE:
|
|
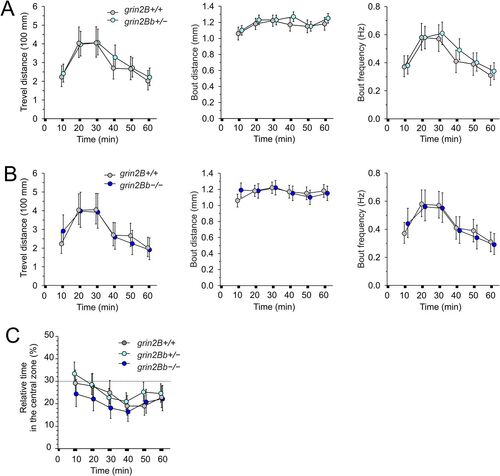
No effect of grin2Bb deletion on larval locomotor activity and thigmotaxis. Comparison of the MTD, MBD, and MBF assessed in grin2Bb+/− (A) and grin2Bb−/− (B) larvae compared with grin2B+/+ larvae (6 dpf). C, Thigmotaxis measured as the relative time spent in the central zone by grin2Bb+/− and grin2Bb−/− larvae compared with grin2B+/+ larvae. The analysis of thigmotaxis was performed on the same larvae used to study locomotion. The dotted line indicates the level of randomness (30%). Data are expressed as mean ± 95% confidence interval of travel distance, bout distance, and bout frequency (A, B) and the relative time spent by larvae in the central zone (C); data were analyzed by ANOVA followed by the post hoc LSD tests (grin2B+/+, n = 198; grin2Bb+/−, n = 288; grin2Bb−/−, n = 144). grin2Bb+/− versus grin2B+/+, travel distance genotype, p = 0.18; time, p < 0.001; bout distance genotype, p = 0.37; time, p < 0.003; bout frequency genotype, p = 0.13; time, p < 0.002; the relative time larvae spent in the central zone genotype, p = 0.23; time, p < 0.002. grin2Bb−/− versus grin2B+/+, travel distance genotype, p = 0.97; time, p < 0.001; bout distance genotype, p = 0.53; time, p = 0.35; bout frequency genotype, p = 0.83; time, p < 0.001; the relative time larvae spent in the central zone genotype, p = 0.11; time, p = 0.23. PHENOTYPE:
|
|
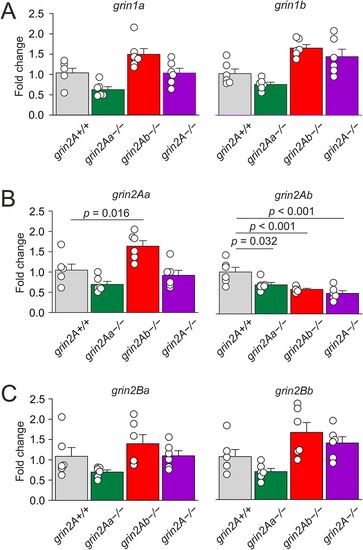
Effects of grin2Aa or grin2Ab deletion on grin mRNA expression in zebrafish larvae. Relative expression levels of grin1 (A), grin2A (B), and grin2B (C) mRNA in heads of zebrafish larvae (6 dpf) for grin2A+/+, grin2Aa−/−, grin2Ab−/−, and grin2A−/− fish were analyzed by RT-qPCR. One-way ANOVA was used to assess the differences in the relative expression of each gene, followed by multiple comparisons versus the corresponding gene in the grin2A+/+ larvae (Dunnett's method), with significance as indicated. grin1a, p = 0.028; grin1b, p = 0.010; grin2Aa, p < 0.001; grin2Ab, p < 0.001; grin2Ba, p = 0.030; grin2Bb, p = 0.011; n = 6 for each genotype, grin2A+/+, grin2Aa−/−, grin2Ab−/−, and grin2A−/−, for each analysis. For effects of grin2Aa or grin2Ab deletion on grin mRNA expression in the adult zebrafish, see Extended Data Figure 7-1. |
|
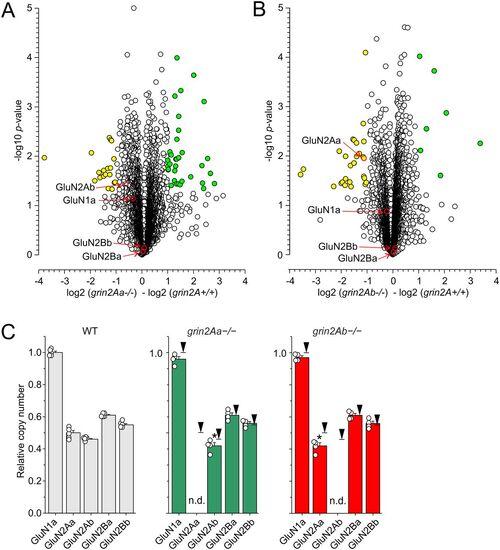
Proteomic analysis of the optic tectum from grin2A+/+, grin2Aa−/−, and grin2Ab−/− adult fish. The volcano plots show proteomics data. The abscissa displays negative (downregulated) and positive (upregulated) fold changes in the ratio of protein levels found in grin2Aa−/− relative to grin2A+/+ (A) and grin2Ab−/− relative to grin2A+/+ (B). The statistical significance (-log of p values; indicated on the ordinate) was assessed using two-sample test with the level of significance determined using permutation-based FDR (see Materials and Methods for details). Yellow symbols show proteins with significantly downregulated expression and green symbols show proteins with significantly upregulated expression (see Extended Data Figs. 8-1, 8-2 for details of proteins with altered expression). C, The plots show the relative CAN of GluN1a, GluN2Aa, GluN2Ab, GluN2Ba, and GluN2Bb expressed in grin2A+/+ (on the left), grin2Aa−/− (in the middle), and grin2Ab−/− fish (on the right). CAN of individual NMDAR subunits was normalized with respect to CAN of GluN1a assessed in the grin2A+/+ fish. Black arrows indicate relative expression in the grin2A+/+ fish; n.d. stands for not detected. One-way ANOVA was used to assess the significance in the relative CAN of NMDAR subunits. Relative CAN of GluN2Aa and GluN2Ab were significantly changed when compared with grin2A+/+ (*). For proteomic analysis of selected presynaptic proteins, see Extended Data Figure 8-3. |