- Title
-
Disruption of the moonlighting function of CTF18 in a patient with T-lymphopenia
- Authors
- Sertori, R., Truong, B., Singh, M.K., Shinton, S., Price, R., Sharo, A., Shultes, P., Sunderam, U., Rana, S., Srinivasan, R., Datta, S., Font-Burgada, J., Brenner, S.E., Puck, J.M., Wiest, D.L.
- Source
- Full text @ Front Immunol
|
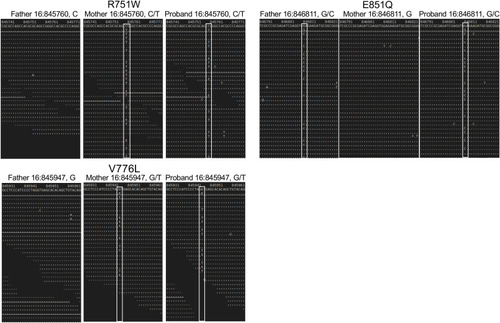
Sequence analysis of the parent and proband |
|
|
|
|
|
|
|
|
|
|
|
|

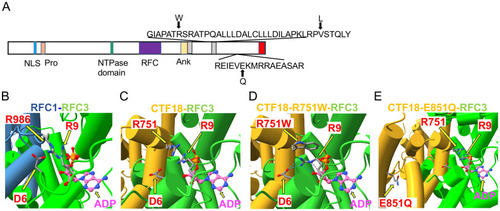
ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

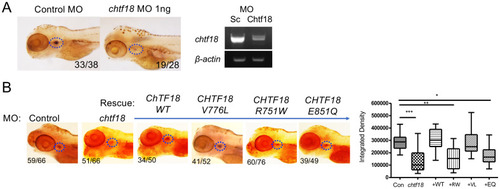
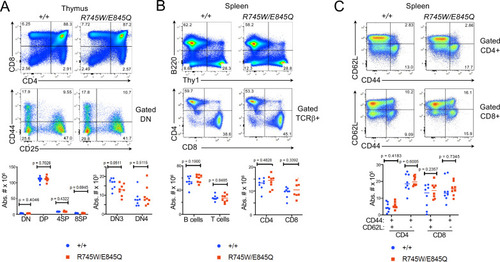
ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |