Fig. 1
- ID
- ZDB-FIG-250626-26
- Publication
- Capps et al., 2025 - Disrupted diencephalon development and neuropeptidergic pathways in zebrafish with autism-risk mutations
- Other Figures
- All Figure Page
- Back to All Figure Page
|
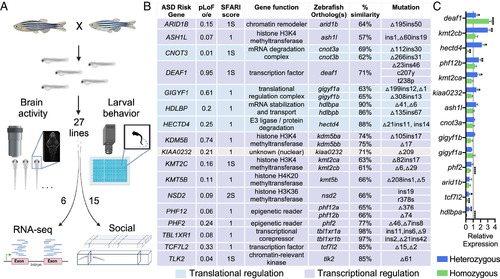
Generation and analysis of mutants for genes that increase risk for ASD. (A) Pipeline of the screen. Mutant lines generated with CRISPR/Cas9 were crossed together, and sibling larvae were assessed for changes to brain morphology, brain activity, and behavioral profiling in 96-well plates. Subsets of lines were further characterized with bulk mRNA-sequencing of dissected heads or social behavior. Created with BioRender.com. (B) Selected genes that increase risk for ASD and corresponding zebrafish CRISPR/Cas9 alleles. The pLoF observed/expected (o/e) score for loss-of-function mutations is from gnomAD database, and a low value indicates the gene is resistant to mutation in humans. A Simons Foundation Autism Research Initiative (SFARI) score of 1 indicates that the genes are implicated in ASD with high confidence, and the syndromic (S) designation indicates genes associated with increased ASD risk and additional characteristics. (C) The relative change to mRNA levels using qPCR for selected zebrafish mutant lines. The heterozygous and homozygous samples are normalized relative to the wild-type level, indicated by the dashed line at 1. Statistical significance represents comparison to the wild-type level, using Brown–Forsythe and Welch ANOVA corrected for multiple comparisons. Data for the remaining mutants are available in SI Appendix, Fig. S3. |