- Title
-
Disrupted development of sensory systems and the cerebellum in a zebrafish ebf3a mutant
- Authors
- Dang, N.D.P., Barcus, A.K., Conklin, C.L., Truong, T.Q., Vivian, M.D., Wang, J., Thomas, H.R., Parant, J.M., Yeo, N.C., Thyme, S.B.
- Source
- Full text @ G3 (Bethesda)
|
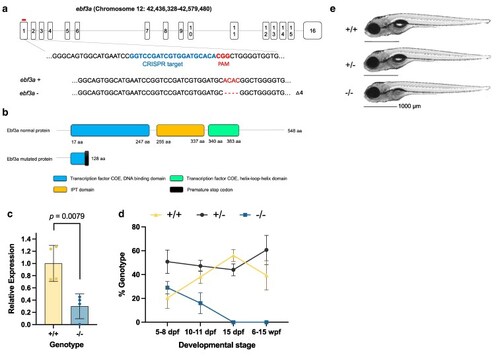
Generation and developmental characterization of zebrafish |
|
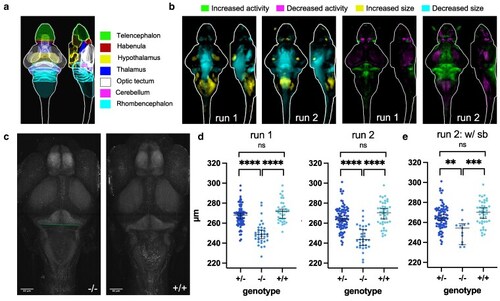
Brain structure and activity phenotypes of zebrafish |
|
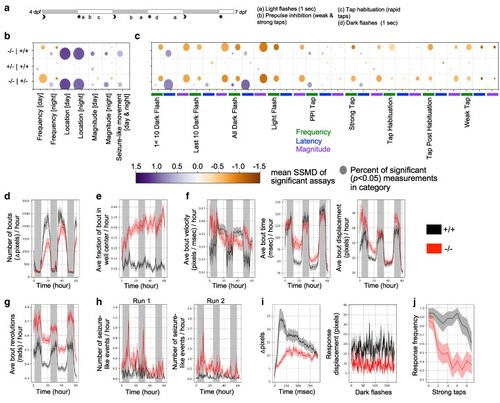
Behavioral phenotypes of zebrafish |
|
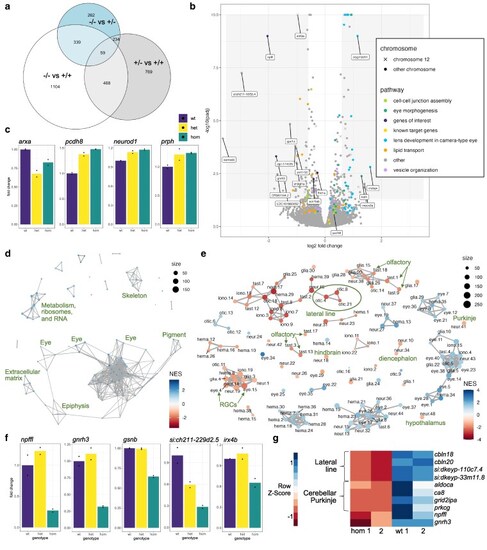
Transcriptional changes in zebrafish |
|
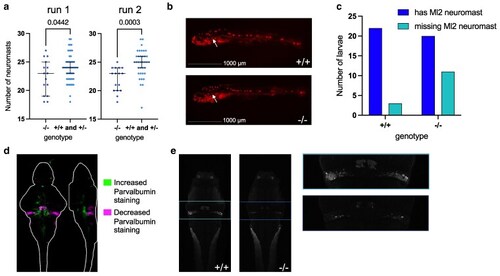
Staining of the lateral line and Purkinje neurons in zebrafish |