- Title
-
Zebrafish adamtsl4 knockout recapitulates key features of human ADAMTSL4-related diseases: a gene involved in extracellular matrix organization, cell junctions and development
- Authors
- Tevar, A., Aroca-Aguilar, J.D., Atiénzar-Aroca, R., Ramírez, A.I., Fernández-Albarral, J.A., Escribano, J.
- Source
- Full text @ Exp. Eye. Res.
|
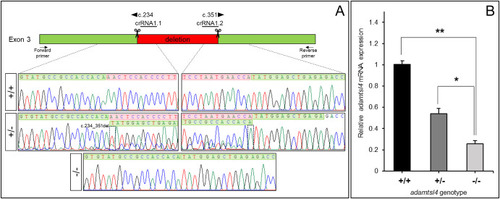
Molecular characterization of the adamtsl4-KO zebrafish line generated using CRISPR/Cas9 genome editing. A. Localization of the deletion identified in adamtsl4 exon 3. Scissors: DNA cleavage sites targeted by two crRNAs (crRNA1.1 and crRNA1.2). The numbers indicate the cDNA nucleotide positions corresponding to the start and end of the deleted DNA fragment. Wild-type sequences are highlighted with a green background, while deleted sequences are marked with a red background. Arrows: position of the forward and reverse primers used for zebrafish genotyping. Rectangles in the electropherograms mark the 5′ and 3′ ends of the deletion. B. Reduced mRNA levels in adamtsl4-KO zebrafish larvae. Relative adamtsl4 mRNA levels were measured in 6 days post-fertilization (dpf) zebrafish larvae using RT-qPCR. Data represent the mean ± standard error, derived from three independent experiments performed in triplicate, using pools of 50 larvae. Statistically significant differences between KO and wild-type zebrafish are denoted by asterisks: p < 0.05 (∗), p < 0.01 (∗∗). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.) |
|
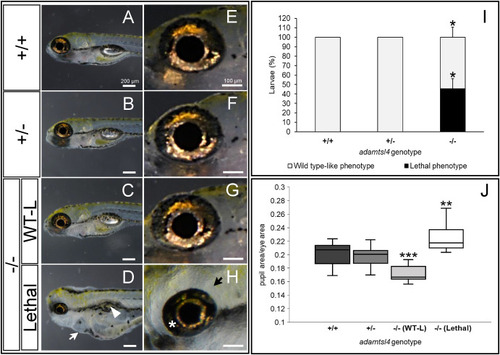
Morphological characterization of F3 adamtsl4-KO larvae at 6 dpf. Representative photographs of wild type (+/+), heterozygous (±), wild type-like knockout (−/− (WT-L)), and lethal knockout (−/− (Lethal)) larvae are shown. A-D. General body morphology. White arrow: pericardial edema; White arrowhead: abnormal swim bladder. E-H. Magnification of ocular structures. I. Quantification of the percentage of KO larvae displaying wild type-like and lethal phenotypes. The data represent three independent replicates with the following total sample sizes: wild type, n = 31; heterozygous, n = 59; mutant homozygous, n = 23. J. Quantification of pupil area normalized to total eye area. The following sample sizes were used for eyes: wild type, n = 24; heterozygous, n = 24; homozygous, n = 24. ∗: p < 0.05; ∗∗: p < 0.01; ∗∗∗: p < 0.001. PHENOTYPE:
|
|
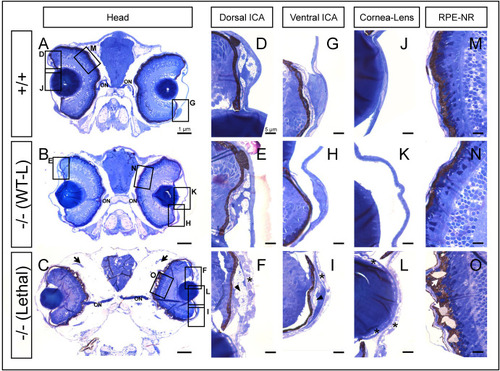
Histological analysis of toluidine blue-stained tissue sections from adamtsl4 KO zebrafish larvae (6 dpf). Transverse semithin tissue sections (500 nm) were prepared as described in the Materials and Methods section. Representative photographs of wild type (+/+), wild type-like KO (−/− (WT-L)) and lethal KO (−/− (Lethal)) larvae are shown. A-C. Tissue sections from the central part of the eyeball (note the presence of the optic nerve (ON)); rectangles indicate areas magnified in subsequent panels. D-F. Dorsal iridocorneal angle (ICA). G-I. Ventral ICA. J-L. Cornea and lens. M-O. Retinal pigmented epithelium (RPE)-neuroretina (NR) interphase. Asterisks: abnormal intercellular separations; black arrows: periocular edema; black arrowhead: increased xantophores; white arrows: amorphous deposits in the RPE. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.) PHENOTYPE:
|
|
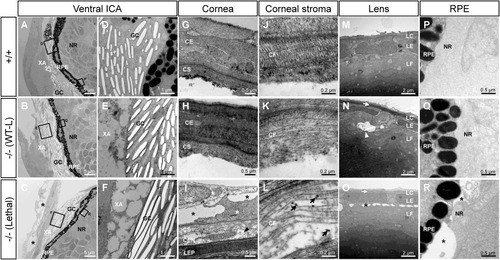
Transmission electron microscopy of the ocular anterior segment in wild type and F3 adamtsl4 KO zebrafish larvae (6 dpf). Tissue sections were prepared as described in the Materials and Methods section. A-C. Representative photographs of wild type (+/+), wild type-like KO (−/− (WT-L)) and lethal KO (−/− (Lethal)) larvae. ICA: ventral iridocorneal angle. The rectangles indicate the areas magnified in subsequent panels. D-F. Ultrastructural details of iridophores and xantophores (XA), showing the presence of guanine crystals (GC) in the iridophores. G-I. Cornea. Note that the corneal endothelium was lost during tissue preparation in panels G and H. J-L. Ultrastructural details of the corneal stroma. M-O. Lens. P-R. Retinal pigment epithelium (RPE)-neuroretina (NR) interphase. CE, corneal epithelium; CEN, corneal endothelium; CF, collagen fibrils; CS, corneal stroma; GC, guanine crystals; LC, lens capsule; LEP, lens epithelium; LF, lens fibers; NR, neuroretina; RPE, retinal pigment epithelium; XA, xanthophores. Black arrows: enlarged and electron-dense CF. Black arrowhead: necrotic CE cell; black asterisks: abnormal intercellular separations; white arrowhead: autophagy; white arrows: thickened LC; white asterisks: swollen necrotic cells. |
|
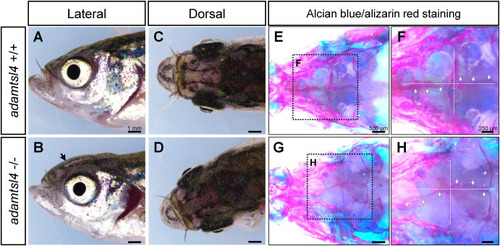
Craniofacial characterization of 5 months F3 adamtsl4-KO zebrafish. A-D. Representative images of wild-type and adamtsl4-KO zebrafish before clearing and staining, highlighting gross craniofacial morphology. E-H. Staining with alcian blue for cartilage and alizarin red for bone structures reveals detailed craniofacial anatomy. Rectangles indicate regions of interest magnified in subsequent panels, providing a closer examination of structural differences between genotypes. White arrows indicate intracranial sutures. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.) PHENOTYPE:
|
|
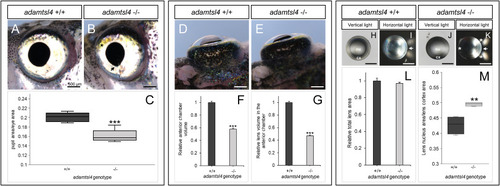
Morphological phenotype characterization of 5 months F3 adamtsl4-KO zebrafish. A-B. Lateral morphology of the eye. (C) Quantification of pupil area relative to the eye area. Sample size: wild-type, n = 5; adamtsl4-KO, n = 9. D-E. Dorsal morphology of the eyes. F. Quantification of anterior chamber volume. G. Quantification of the visible lens volume within the anterior chamber. Sample size in F–G: wild-type, n = 20; adamtsl4-KO, n = 72. H-K. Lens morphology and light transmission. L. Quantification of total lens area. M. Quantification of lens nucleus area relative to the cortex area. Sample sizes: wild-type, n = 4; adamtsl4-KO, n = . ∗∗: p < 0.01; ∗∗∗: p < 0.001. Black asterisk: increased xanthophores. Black arrow: irregular pupil morphology. White asterisk: impaired light transmission in the lens. White arrow: direction of light. Cx, cortex; nu, nucleus. PHENOTYPE:
|
|
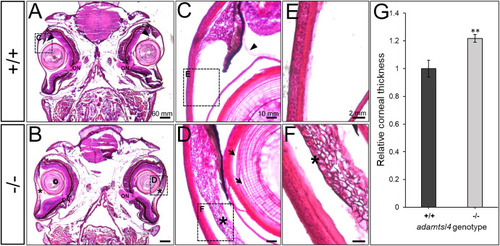
Histological analysis of hematoxylin-eosin-stained tissue sections from adult adamtsl4 KO zebrafish (5 months). Transverse tissue sections were prepared as described in the Materials and Methods section. Representative photographs of wild type (+/+) and KO (−/−) zebrafish are shown. A-B. Tissue sections from the central part of the eyeball highlighting the presence of the optic nerve (ON). C-D. Anterior segment. E-F. Corneal structure. G. Quantitative analysis of corneal thickness. ∗∗: p < 0.01. Rectangles indicate areas magnified in subsequent panels. Arrows: separation of lens fibers; Asterisks: abnormal annular ligament. PHENOTYPE:
|
|
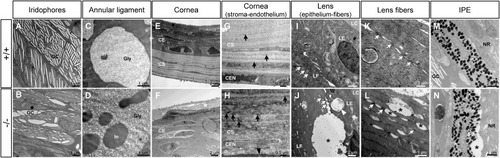
Transmission electron microscopy of the ocular anterior segment structures in adult (3-months-old) F3 adamtsl4 KO zebrafish. Tissue sections were prepared as described in the Materials and Methods section. Representative images of wild type (+/+) and KO (−/−) zebrafish are shown. A-B. Guanine crystals (GC) in iridophores. C-D. Glycoprotein aggregates (Gly) and cytoplasmic inclusions (In) in annular ligament cells. E-F. Cornea showing the thickened corneal epithelium (CE) and the necrotic superficial epithelial cell (white asterisk) in the KO zebrafish. G-H. Corneal stroma (CS) and endothelium (CEN). Note the contact between the CEN with the abnormal annular ligament in the KO zebrafish (black arrowhead). I-J. Lens epithelium (LE)-fiber (LF) interphase. K-L. Lens fibers. Gap junctions (white arrows). M-N. Iris pigment epithelium (IPE)-neuroretina (NE) interphase. Black asterisks: abnormal intercellular separations. |
|
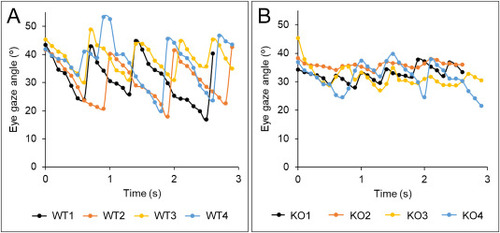
Optokinetic response in adult zebrafish at 8 months based on frame-by-frame eye angle measurements. A. Wild-type zebrafish. B. adamtsl4-KO zebrafish. Films were analyzed using Image-Pro Plus software to determine eye angles across individual frames. Data points represent eye angles measured in a representative analysis of four fish per group. PHENOTYPE:
|
|
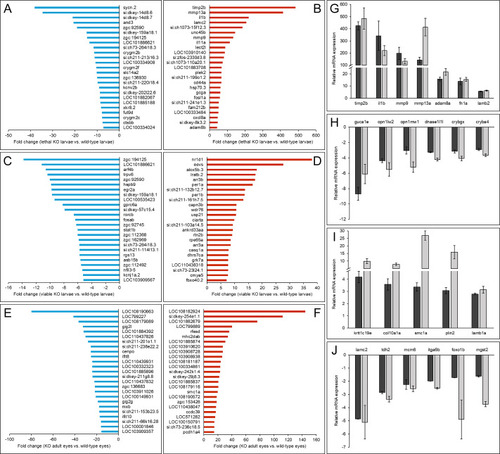
Top 50 DEGs in adamtsl4 KO zebrafish identified by RNA-seq analysis and confirmation by RT-qPCR of the differential expression of selected genes. A-B. Downregulated genes and upregulated genes, respectively, in 6 dpf lethal adamtsl4 KO larvae vs. wild type larvae. C-D. Downregulated genes and upregulated genes, respectively, in 6 dpf wild type-like adamtsl4 KO larvae vs. wild type larvae. E-F. Downregulated genes and upregulated genes, respectively, in eyes of 5-month adamtsl4 KO zebrafish vs. wild type zebrafish. G-H. Confirmation by qRT-PCR of differential gene expression of selected upregulated and downregulated genes, respectively, in lethal adamtsl4 KO larvae vs. wild type larvae. I-J. Confirmation by qRT-PCR of differential gene expression of selected upregulated and downregulated genes, respectively, in eyes of 5-monts adamtsl4 KO zebrafish vs. wild type zebrafish. Aliquots of RNA preparations used for transcriptomic analyses were used as templates in qRT-PCR that was carried out in triplicate. |
Reprinted from Experimental Eye Research, , Tevar, A., Aroca-Aguilar, J.D., Atiénzar-Aroca, R., Ramírez, A.I., Fernández-Albarral, J.A., Escribano, J., Zebrafish adamtsl4 knockout recapitulates key features of human ADAMTSL4-related diseases: a gene involved in extracellular matrix organization, cell junctions and development, 110572110572, Copyright (2025) with permission from Elsevier. Full text @ Exp. Eye. Res.