- Title
-
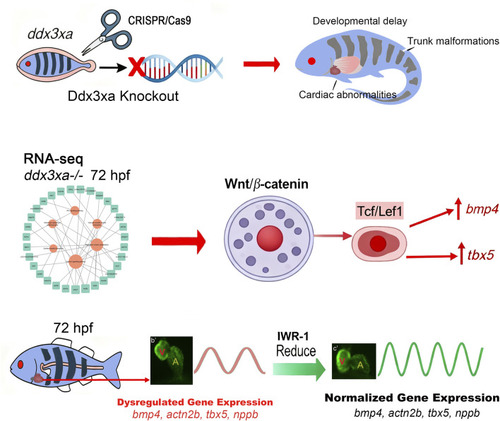
Ddx3xa mutations drive cardiac defects in a zebrafish model via dysregulation of wnt/β-catenin signaling
- Authors
- Chen, Y., Lin, M., Zhu, P., Wang, H., Jiao, Z., Yi, K., Yang, X., Zhang, Y., Cai, X., Yuan, W., Li, Y., Jiang, Z., Wang, Y., Li, F., Wu, X., Fan, X.
- Source
- Full text @ Front Mol Biosci
|
Phylogenetic tree and amino acid sequence analysis of DDX3X across species. |
|
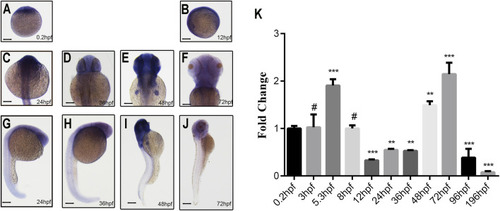
Spatiotemporal expression pattern of |
|
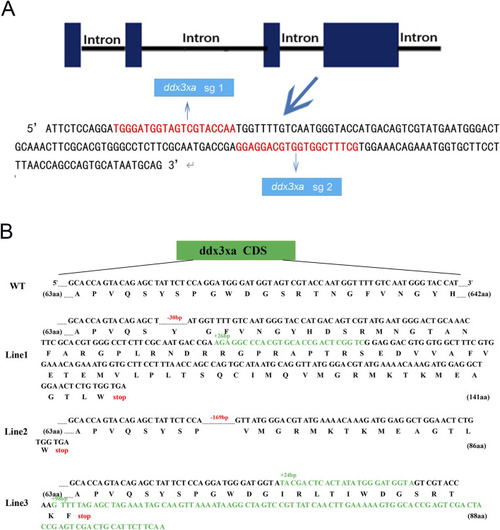
Schematic representation of |
|
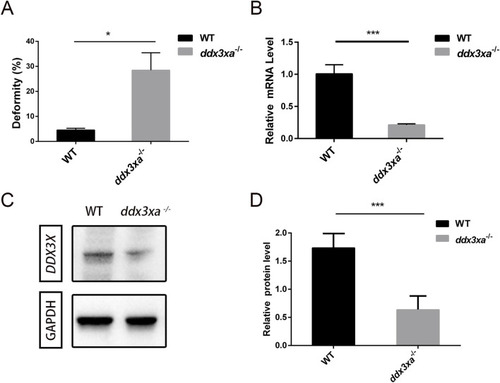
Malformation rate quantification and validation of EXPRESSION / LABELING:
PHENOTYPE:
|
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
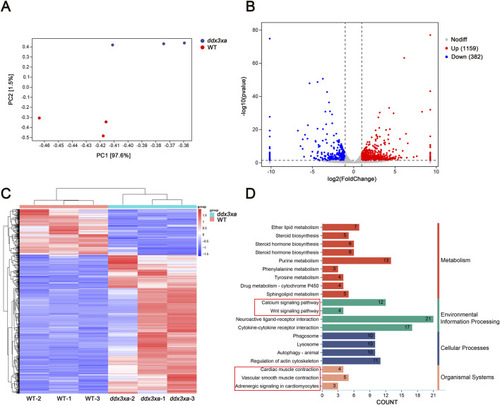
Transcriptomic alterations in |
|
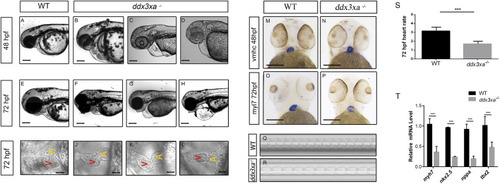
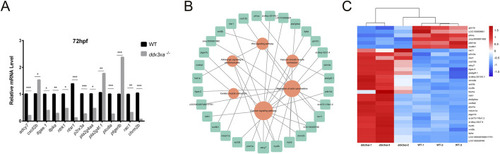
Validation and pathway analysis of cardiac development genes in EXPRESSION / LABELING:
PHENOTYPE:
|
|
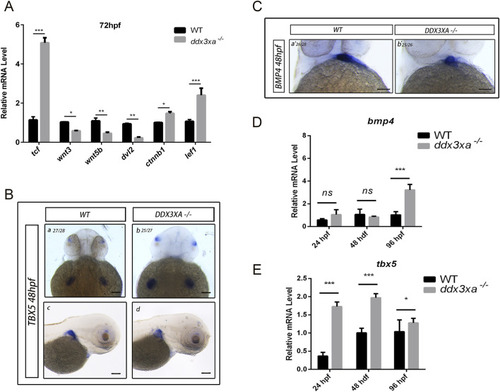
Wnt signaling disruption in |
|
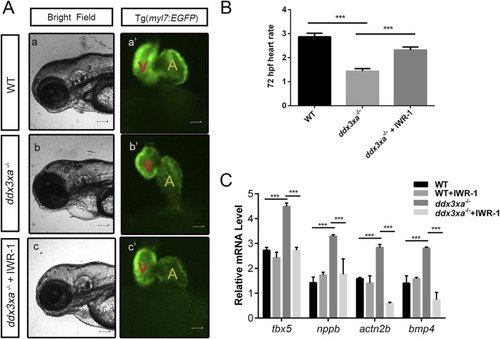
Pharmacological rescue of cardiac defects by Wnt inhibition. EXPRESSION / LABELING:
PHENOTYPE:
|
|
|